CRISPR-U™ Gene Knockout Cell Line
Strategy
DEPDC5 Gene Knockout Strategy
CRISPR-U™ technology (CRISPR based), developed by Ubigene, is more efficient than general
CRISPR/Cas9 technology in double-strand breaking and homologous recombination. With CRISPR-U™, Ubigene has
successfully edited over 3000 genes on more than 100 types of cell lines.
Objective
To create a Human DEPDC5 Knockout
model in cell line by
CRISPR-U™-mediated
genome
engineering.
Target gene info 
| Official symbol | DEPDC5 |
| Gene id | 9681 |
| Organism | Homo sapiens |
| Official full symbol | DEP domain containing 5, GATOR1 subcomplex subunit |
| Gene type | protein-coding |
| Also known as | DEP.5, FFEVF, FFEVF1 |
| Summary | This gene encodes a member of the IML1 family of proteins involved in G-protein signaling pathways. The mechanistic target of rapamycin complex 1 (mTORC1) pathway regulates cell growth by sensing the availability of nutrients. The protein encoded by this gene is a component of the GATOR1 (GAP activity toward Rags) complex which inhibits the amino acid-sensing branch of the mTORC1 pathway. Mutations in this gene are associated with autosomal dominant familial focal epilepsy with variable foci. A single nucleotide polymorphism in an intron of this gene has been associated with an increased risk of hepatocellular carcinoma in individuals with chronic hepatitis C virus infection. Alternative splicing results in multiple transcript variants. |
| Genomic regions | Chromosome 22 |
Strategy Summary
This gene has 27 protein coding transcripts:
| Name | Transcript ID | bp | Protein | Biotype | CCDS | UniProt Match | RefSeq Match | Flags |
| DEPDC5-264 | ENST00000651528.2 | 6554 | 1603aa | Protein coding | CCDS74849 | O75140-10 | NM_001242896.3 | GENCODE basic, APPRIS P4, MANE Select v0.92, |
| DEPDC5-226 | ENST00000642696.1 | 6479 | 1572aa | Protein coding | CCDS43006 | O75140-1 | - | GENCODE basic, APPRIS ALT1, |
| DEPDC5-248 | ENST00000645711.1 | 5450 | 1594aa | Protein coding | CCDS46692 | O75140-9 | - | GENCODE basic, APPRIS ALT1, |
| DEPDC5-202 | ENST00000382112.8 | 5390 | 1603aa | Protein coding | CCDS74849 | O75140-10 | - | TSL:1, GENCODE basic, APPRIS P4, |
| DEPDC5-239 | ENST00000644331.1 | 5383 | 1581aa | Protein coding | CCDS87017 | O75140-4 | - | GENCODE basic, APPRIS ALT1, |
| DEPDC5-206 | ENST00000400249.7 | 5350 | 1581aa | Protein coding | CCDS87017 | O75140-4 | - | TSL:5, GENCODE basic, APPRIS ALT1, |
| DEPDC5-205 | ENST00000400248.7 | 5319 | 1572aa | Protein coding | CCDS43006 | O75140-1 | - | TSL:1, GENCODE basic, APPRIS ALT1, |
| DEPDC5-259 | ENST00000646969.1 | 5286 | 1525aa | Protein coding | CCDS87018 | A0A2R8Y6H3 | - | GENCODE basic, |
| DEPDC5-245 | ENST00000645560.1 | 5207 | 1525aa | Protein coding | CCDS87018 | A0A2R8Y6H3 | - | GENCODE basic, |
| DEPDC5-221 | ENST00000535622.6 | 5197 | 1503aa | Protein coding | CCDS56229 | O75140-8 | - | TSL:2, GENCODE basic, |
| DEPDC5-203 | ENST00000400242.8 | 2448 | 559aa | Protein coding | CCDS43007 | O75140-2 | - | TSL:1, GENCODE basic, |
| DEPDC5-257 | ENST00000646755.1 | 2317 | 559aa | Protein coding | CCDS43007 | O75140-2 | - | GENCODE basic, |
| DEPDC5-207 | ENST00000433147.2 | 5331 | 1575aa | Protein coding | - | H0Y770 | - | TSL:1, GENCODE basic, |
| DEPDC5-255 | ENST00000646465.1 | 5258 | 1496aa | Protein coding | - | A0A2R8Y721 | - | GENCODE basic, |
| DEPDC5-201 | ENST00000382111.6 | 5212 | 1552aa | Protein coding | - | O75140-5 | - | TSL:5, GENCODE basic, |
| DEPDC5-242 | ENST00000645407.1 | 5010 | 1589aa | Protein coding | - | A0A2R8Y7U0 | - | GENCODE basic, |
| DEPDC5-262 | ENST00000647343.1 | 4664 | 1484aa | Protein coding | - | A0A2R8Y5K9 | - | CDS 3' incomplete, |
| DEPDC5-230 | ENST00000642974.1 | 2682 | 829aa | Protein coding | - | A0A2R8Y7U6 | - | GENCODE basic, |
| DEPDC5-246 | ENST00000645564.1 | 2542 | 795aa | Protein coding | - | A0A2R8Y5E9 | - | CDS 3' incomplete, |
| DEPDC5-247 | ENST00000645693.1 | 2403 | 748aa | Protein coding | - | A0A2R8YF97 | - | CDS 3' incomplete, |
| DEPDC5-263 | ENST00000647438.1 | 2221 | 531aa | Protein coding | - | A0A2R8Y5P2 | - | GENCODE basic, |
| DEPDC5-208 | ENST00000437411.6 | 1478 | 193aa | Protein coding | - | C9JGS4 | - | TSL:5, GENCODE basic, |
| DEPDC5-241 | ENST00000645015.1 | 1278 | 193aa | Protein coding | - | C9JGS4 | - | GENCODE basic, |
| DEPDC5-211 | ENST00000458532.2 | 914 | 135aa | Protein coding | - | H7C3I3 | - | CDS 5' incomplete, TSL:5, |
| DEPDC5-210 | ENST00000456178.6 | 549 | 37aa | Protein coding | - | F8WAX3 | - | CDS 3' incomplete, TSL:5, |
| DEPDC5-216 | ENST00000473802.1 | 503 | 48aa | Protein coding | - | A0A2R8YEI8 | - | CDS 3' incomplete, TSL:3, |
| DEPDC5-220 | ENST00000497340.1 | 288 | 30aa | Protein coding | - | A0A2R8Y7X0 | - | CDS 5' incomplete, TSL:5, |
| DEPDC5-204 | ENST00000400246.7 | 5725 | 1343aa | Nonsense mediated decay | - | A0A5F9UWT1 | - | TSL:1, |
| DEPDC5-256 | ENST00000646515.1 | 5472 | 1198aa | Nonsense mediated decay | - | A0A2R8Y5T1 | - | - |
| DEPDC5-225 | ENST00000642684.1 | 5410 | 95aa | Nonsense mediated decay | - | A0A2R8YET0 | - | - |
| DEPDC5-243 | ENST00000645494.1 | 5371 | 638aa | Nonsense mediated decay | - | A0A2R8Y6H8 | - | - |
| DEPDC5-260 | ENST00000646998.1 | 5302 | 1285aa | Nonsense mediated decay | - | A0A2R8YEW8 | - | - |
| DEPDC5-227 | ENST00000642771.1 | 5301 | 638aa | Nonsense mediated decay | - | A0A2R8Y6H8 | - | - |
| DEPDC5-238 | ENST00000644162.1 | 5224 | 405aa | Nonsense mediated decay | - | A0A2R8YFS1 | - | - |
| DEPDC5-249 | ENST00000645755.1 | 5214 | 333aa | Nonsense mediated decay | - | A0A2R8Y6Y2 | - | - |
| DEPDC5-236 | ENST00000643751.2 | 5067 | 1187aa | Nonsense mediated decay | - | A0A2R8Y6V4 | - | - |
| DEPDC5-235 | ENST00000643395.1 | 4790 | 1207aa | Nonsense mediated decay | - | A0A2R8Y842 | - | - |
| DEPDC5-209 | ENST00000448753.6 | 3256 | 659aa | Nonsense mediated decay | - | H7C1T0 | - | CDS 5' incomplete, TSL:1, |
| DEPDC5-244 | ENST00000645547.1 | 2084 | 228aa | Nonsense mediated decay | - | A0A2R8Y7C9 | - | CDS 5' incomplete, |
| DEPDC5-252 | ENST00000645967.1 | 1675 | 153aa | Nonsense mediated decay | - | A0A2R8Y6F4 | - | - |
| DEPDC5-214 | ENST00000469974.6 | 880 | 116aa | Nonsense mediated decay | - | A0A2R8Y5V3 | - | CDS 5' incomplete, TSL:5, |
| DEPDC5-231 | ENST00000643021.1 | 2249 | No protein | Processed transcript | - | - | - | - |
| DEPDC5-233 | ENST00000643097.1 | 2211 | No protein | Processed transcript | - | - | - | - |
| DEPDC5-240 | ENST00000644690.1 | 2052 | No protein | Processed transcript | - | - | - | - |
| DEPDC5-258 | ENST00000646830.1 | 1518 | No protein | Processed transcript | - | - | - | - |
| DEPDC5-219 | ENST00000494060.1 | 750 | No protein | Processed transcript | - | - | - | TSL:5, |
| DEPDC5-223 | ENST00000642551.1 | 4516 | No protein | Retained intron | - | - | - | - |
| DEPDC5-251 | ENST00000645893.1 | 3375 | No protein | Retained intron | - | - | - | - |
| DEPDC5-253 | ENST00000646135.1 | 2776 | No protein | Retained intron | - | - | - | - |
| DEPDC5-222 | ENST00000642212.1 | 2624 | No protein | Retained intron | - | - | - | - |
| DEPDC5-234 | ENST00000643166.1 | 2300 | No protein | Retained intron | - | - | - | - |
| DEPDC5-217 | ENST00000479261.2 | 2173 | No protein | Retained intron | - | - | - | TSL:2, |
| DEPDC5-254 | ENST00000646383.1 | 2160 | No protein | Retained intron | - | - | - | - |
| DEPDC5-261 | ENST00000647289.1 | 2112 | No protein | Retained intron | - | - | - | - |
| DEPDC5-228 | ENST00000642915.1 | 2064 | No protein | Retained intron | - | - | - | - |
| DEPDC5-237 | ENST00000643948.1 | 1424 | No protein | Retained intron | - | - | - | - |
| DEPDC5-229 | ENST00000642956.1 | 1397 | No protein | Retained intron | - | - | - | - |
| DEPDC5-224 | ENST00000642605.1 | 741 | No protein | Retained intron | - | - | - | - |
| DEPDC5-213 | ENST00000469969.2 | 679 | No protein | Retained intron | - | - | - | TSL:5, |
| DEPDC5-218 | ENST00000490731.1 | 602 | No protein | Retained intron | - | - | - | TSL:4, |
| DEPDC5-250 | ENST00000645785.1 | 584 | No protein | Retained intron | - | - | - | - |
| DEPDC5-215 | ENST00000471914.5 | 569 | No protein | Retained intron | - | - | - | TSL:4, |
| DEPDC5-232 | ENST00000643075.1 | 356 | No protein | Retained intron | - | - | - | - |
| DEPDC5-212 | ENST00000462414.1 | 231 | No protein | Retained intron | - | - | - | TSL:5, |




Red Cotton™ Assessment
Project Difficulty Level unknown
| Target Gene | DEPDC5 |
| This KO Strategy | loading |
| Red Cotton™ Notes | Gene DEPDC5 had been KO in a549 cell line. |
Aforementioned information comes from Ubigene database. Different origin of cell lines may have
different condition. Ubigene reserved all the right for final explanation.
Special deals for this gene:
$49
Single gRNA plasmid off-shelf
$599
Single gRNA lentivirus



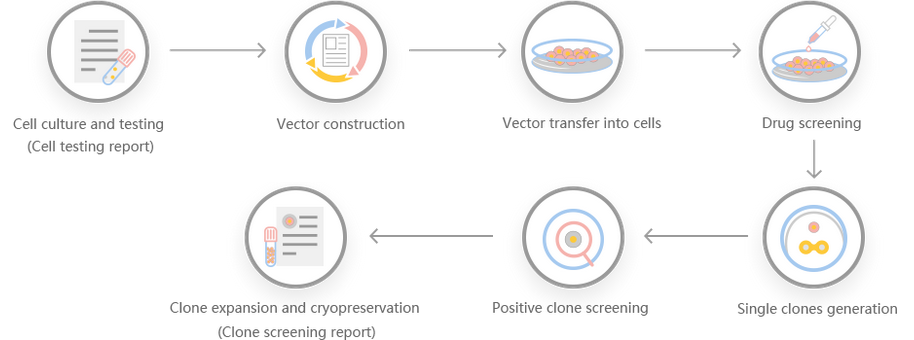
Work flow






 Comment:
Comment: