CRISPR-U™ Gene Knockout Cell Line
Strategy
CAST Gene Knockout Strategy
CRISPR-U™ technology (CRISPR based), developed by Ubigene, is more efficient than general
CRISPR/Cas9 technology in double-strand breaking and homologous recombination. With CRISPR-U™, Ubigene has
successfully edited over 3000 genes on more than 100 types of cell lines.
Objective
To create a Human CAST Knockout
model in cell line by
CRISPR-U™-mediated
genome
engineering.
Target gene info 
| Official symbol | CAST |
| Gene id | 831 |
| Organism | Homo sapiens |
| Official full symbol | calpastatin |
| Gene type | protein-coding |
| Also known as | BS-17, PLACK |
| Summary | The protein encoded by this gene is an endogenous calpain (calcium-dependent cysteine protease) inhibitor. It consists of an N-terminal domain L and four repetitive calpain-inhibition domains (domains 1-4), and it is involved in the proteolysis of amyloid precursor protein. The calpain/calpastatin system is involved in numerous membrane fusion events, such as neural vesicle exocytosis and platelet and red-cell aggregation. The encoded protein is also thought to affect the expression levels of genes encoding structural or regulatory proteins. Alternatively spliced transcript variants encoding different isoforms have been described. |
| Genomic regions | Chromosome 5 |
Strategy Summary
This gene has 38 protein coding transcripts:
| Name | Transcript ID | bp | Protein | Biotype | CCDS | UniProt Match | RefSeq Match | Flags |
| CAST-206 | ENST00000395812.6 | 4506 | 750aa | Protein coding | CCDS54882 | P20810-9 | - | TSL:5, GENCODE basic, APPRIS P4, |
| CAST-247 | ENST00000674984.1 | 4330 | 769aa | Protein coding | CCDS43344 | - | - | GENCODE basic, APPRIS ALT2, |
| CAST-201 | ENST00000309190.9 | 4282 | 686aa | Protein coding | CCDS4082 | P20810-8 | - | TSL:1, GENCODE basic, APPRIS ALT2, |
| CAST-203 | ENST00000338252.7 | 2972 | 695aa | Protein coding | CCDS54883 | P20810-2 | - | TSL:1, GENCODE basic, |
| CAST-213 | ENST00000504465.5 | 2336 | 636aa | Protein coding | CCDS75279 | E9PDE4 | - | TSL:2, GENCODE basic, APPRIS ALT2, |
| CAST-228 | ENST00000509903.5 | 2171 | 673aa | Protein coding | CCDS83017 | E9PCH5 | - | TSL:1, GENCODE basic, |
| CAST-250 | ENST00000675179.1 | 4490 | 791aa | Protein coding | - | - | NM_001750.7 | GENCODE basic, APPRIS ALT2, MANE Select v0.92, |
| CAST-207 | ENST00000395813.5 | 4356 | 708aa | Protein coding | - | P20810-1 | - | TSL:5, GENCODE basic, APPRIS ALT2, |
| CAST-248 | ENST00000675033.1 | 4296 | 667aa | Protein coding | - | - | - | GENCODE basic, APPRIS ALT2, |
| CAST-257 | ENST00000675663.1 | 4263 | 754aa | Protein coding | - | - | - | GENCODE basic, APPRIS ALT2, |
| CAST-246 | ENST00000674702.1 | 4067 | 704aa | Protein coding | - | - | - | GENCODE basic, APPRIS ALT2, |
| CAST-245 | ENST00000674587.1 | 4024 | 609aa | Protein coding | - | - | - | GENCODE basic, |
| CAST-255 | ENST00000675479.1 | 3991 | 662aa | Protein coding | - | - | - | GENCODE basic, |
| CAST-209 | ENST00000437034.6 | 3377 | 436aa | Protein coding | - | H0Y7F0 | - | CDS 5' incomplete, TSL:1, |
| CAST-202 | ENST00000325674.11 | 3356 | 430aa | Protein coding | - | A0A0A0MR45 | - | TSL:5, GENCODE basic, |
| CAST-234 | ENST00000511049.5 | 3273 | 693aa | Protein coding | - | E7EVY3 | - | TSL:5, GENCODE basic, APPRIS ALT2, |
| CAST-251 | ENST00000675185.1 | 2772 | 252aa | Protein coding | - | - | - | CDS 5' incomplete, |
| CAST-256 | ENST00000675614.1 | 2727 | 681aa | Protein coding | - | - | - | GENCODE basic, |
| CAST-223 | ENST00000508608.6 | 2699 | 741aa | Protein coding | - | A0A0C4DGB5 | - | TSL:2, GENCODE basic, |
| CAST-224 | ENST00000508830.5 | 2539 | 791aa | Protein coding | - | P20810-6 | - | TSL:5, GENCODE basic, APPRIS ALT2, |
| CAST-259 | ENST00000675858.1 | 2529 | 615aa | Protein coding | - | - | - | GENCODE basic, |
| CAST-204 | ENST00000341926.7 | 2487 | 708aa | Protein coding | - | P20810-1 | - | TSL:1, GENCODE basic, APPRIS ALT2, |
| CAST-236 | ENST00000511782.5 | 2466 | 694aa | Protein coding | - | B7Z574 | - | TSL:2, GENCODE basic, APPRIS ALT2, |
| CAST-233 | ENST00000510756.6 | 2428 | 667aa | Protein coding | - | P20810-10 | - | TSL:5, GENCODE basic, APPRIS ALT2, |
| CAST-230 | ENST00000510156.5 | 2079 | 642aa | Protein coding | - | E7ES10 | - | CDS 3' incomplete, TSL:5, |
| CAST-232 | ENST00000510500.5 | 1835 | 465aa | Protein coding | - | H0Y9H6 | - | CDS 5' incomplete, TSL:1, |
| CAST-222 | ENST00000508579.5 | 1758 | 423aa | Protein coding | - | E7EQ12 | - | TSL:2, GENCODE basic, |
| CAST-244 | ENST00000515663.5 | 1473 | 431aa | Protein coding | - | E7EQA0 | - | TSL:2, GENCODE basic, |
| CAST-235 | ENST00000511097.6 | 1399 | 393aa | Protein coding | - | F8W7E0 | - | CDS 3' incomplete, TSL:5, |
| CAST-208 | ENST00000421689.6 | 913 | 304aa | Protein coding | - | E9PSG1 | - | CDS 3' incomplete, TSL:5, |
| CAST-238 | ENST00000512620.5 | 904 | 301aa | Protein coding | - | H0YA91 | - | CDS 5' and 3' incomplete, TSL:5, |
| CAST-215 | ENST00000505143.5 | 864 | 288aa | Protein coding | - | H0Y944 | - | CDS 5' and 3' incomplete, TSL:3, |
| CAST-241 | ENST00000514845.5 | 588 | 150aa | Protein coding | - | D6RBR1 | - | CDS 3' incomplete, TSL:4, |
| CAST-217 | ENST00000506811.5 | 582 | 156aa | Protein coding | - | E7EN75 | - | CDS 3' incomplete, TSL:5, |
| CAST-221 | ENST00000508197.1 | 567 | 156aa | Protein coding | - | E7EQK6 | - | CDS 3' incomplete, TSL:3, |
| CAST-225 | ENST00000509259.5 | 565 | 91aa | Protein coding | - | D6RC54 | - | CDS 3' incomplete, TSL:4, |
| CAST-240 | ENST00000514055.5 | 559 | 149aa | Protein coding | - | D6RGF7 | - | CDS 3' incomplete, TSL:5, |
| CAST-211 | ENST00000503828.5 | 499 | 78aa | Protein coding | - | D6RAA8 | - | CDS 3' incomplete, TSL:2, |
| CAST-252 | ENST00000675266.1 | 4323 | 81aa | Nonsense mediated decay | - | - | - | CDS 5' incomplete, |
| CAST-253 | ENST00000675267.1 | 4300 | 110aa | Nonsense mediated decay | - | - | - | - |
| CAST-210 | ENST00000484552.6 | 1767 | 430aa | Nonsense mediated decay | - | H0YD33 | - | TSL:1, |
| CAST-254 | ENST00000675275.1 | 1399 | 94aa | Nonsense mediated decay | - | - | - | CDS 5' incomplete, |
| CAST-229 | ENST00000510098.1 | 1323 | 203aa | Nonsense mediated decay | - | A0A0C4DGD1 | - | TSL:1, |
| CAST-220 | ENST00000508117.6 | 571 | 82aa | Nonsense mediated decay | - | D6RBZ8 | - | TSL:5, |
| CAST-205 | ENST00000348386.7 | 2272 | No protein | Processed transcript | - | - | - | TSL:1, |
| CAST-258 | ENST00000675734.1 | 1111 | No protein | Processed transcript | - | - | - | - |
| CAST-212 | ENST00000504221.5 | 578 | No protein | Processed transcript | - | - | - | TSL:3, |
| CAST-227 | ENST00000509602.1 | 557 | No protein | Processed transcript | - | - | - | TSL:4, |
| CAST-219 | ENST00000507836.1 | 449 | No protein | Processed transcript | - | - | - | TSL:3, |
| CAST-218 | ENST00000507487.5 | 396 | No protein | Processed transcript | - | - | - | TSL:5, |
| CAST-216 | ENST00000505402.1 | 362 | No protein | Processed transcript | - | - | - | TSL:4, |
| CAST-249 | ENST00000675107.1 | 3117 | No protein | Retained intron | - | - | - | - |
| CAST-226 | ENST00000509529.1 | 3066 | No protein | Retained intron | - | - | - | TSL:2, |
| CAST-239 | ENST00000513666.5 | 1539 | No protein | Retained intron | - | - | - | TSL:2, |
| CAST-214 | ENST00000504532.1 | 714 | No protein | Retained intron | - | - | - | TSL:2, |
| CAST-242 | ENST00000515063.1 | 654 | No protein | Retained intron | - | - | - | TSL:1, |
| CAST-243 | ENST00000515160.5 | 617 | No protein | Retained intron | - | - | - | TSL:2, |
| CAST-231 | ENST00000510245.1 | 561 | No protein | Retained intron | - | - | - | TSL:4, |
| CAST-237 | ENST00000512191.1 | 406 | No protein | Retained intron | - | - | - | TSL:2, |




Strategy
Click to get 
Red Cotton™ Assessment
Project Difficulty Level unknown
| Target Gene | CAST |
| This KO Strategy | loading |
| Red Cotton™ Notes | Gene CAST had been KO in hela cell line. |
Aforementioned information comes from Ubigene database. Different origin of cell lines may have
different condition. Ubigene reserved all the right for final explanation.
Special deals for this gene:
$49
Single gRNA plasmid off-shelf
$599
Single gRNA lentivirus



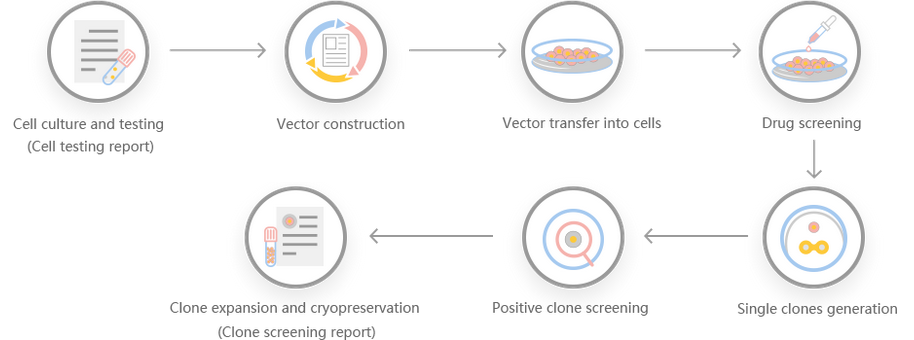
Work flow





 Comment:
Comment: