CRISPR-U™ Gene Knockout Cell Line
Strategy
PTK2 Gene Knockout Strategy
CRISPR-U™ technology (CRISPR based), developed by Ubigene, is more efficient than general
CRISPR/Cas9 technology in double-strand breaking and homologous recombination. With CRISPR-U™, Ubigene has
successfully edited over 3000 genes on more than 100 types of cell lines.
Objective
To create a Human PTK2 Knockout
model in cell line by
CRISPR-U™-mediated
genome
engineering.
Target gene info 
| Official symbol | PTK2 |
| Gene id | 5747 |
| Organism | Homo sapiens |
| Official full symbol | protein tyrosine kinase 2 |
| Gene type | protein-coding |
| Also known as | FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK |
| Summary | This gene encodes a cytoplasmic protein tyrosine kinase which is found concentrated in the focal adhesions that form between cells growing in the presence of extracellular matrix constituents. The encoded protein is a member of the FAK subfamily of protein tyrosine kinases but lacks significant sequence similarity to kinases from other subfamilies. Activation of this gene may be an important early step in cell growth and intracellular signal transduction pathways triggered in response to certain neural peptides or to cell interactions with the extracellular matrix. Several transcript variants encoding different isoforms have been found for this gene. |
| Genomic regions | Chromosome 8 |
Strategy Summary
This gene has 33 protein coding transcripts:
| Name | Transcript ID | bp | Protein | Biotype | CCDS | UniProt Match | RefSeq Match | Flags |
| PTK2-240 | ENST00000522684.5 | 4955 | 1052aa | Protein coding | CCDS6381 | Q05397-1 | - | TSL:1, GENCODE basic, APPRIS P2, |
| PTK2-201 | ENST00000340930.7 | 4414 | 1065aa | Protein coding | CCDS56557 | Q05397-5 | - | TSL:2, GENCODE basic, APPRIS ALT2, |
| PTK2-228 | ENST00000521059.5 | 4405 | 1052aa | Protein coding | CCDS6381 | Q05397-1 | - | TSL:1, GENCODE basic, APPRIS P2, |
| PTK2-203 | ENST00000430260.6 | 1940 | 362aa | Protein coding | CCDS87628 | B4DWJ1 | - | TSL:2, GENCODE basic, |
| PTK2-207 | ENST00000517887.5 | 4661 | 1096aa | Protein coding | - | E7ESA6 | - | TSL:5, GENCODE basic, APPRIS ALT1, |
| PTK2-212 | ENST00000519419.5 | 4439 | 1096aa | Protein coding | - | E7ESA6 | - | TSL:5, GENCODE basic, APPRIS ALT1, |
| PTK2-202 | ENST00000395218.3 | 4415 | 1006aa | Protein coding | - | Q05397-7 | - | TSL:5, GENCODE basic, APPRIS ALT2, |
| PTK2-215 | ENST00000519654.5 | 4129 | 1017aa | Protein coding | - | H0YBP1 | - | CDS 5' incomplete, TSL:1, |
| PTK2-206 | ENST00000517712.5 | 3351 | 115aa | Protein coding | - | E5RHD8 | - | TSL:2, GENCODE basic, |
| PTK2-213 | ENST00000519465.5 | 3279 | 680aa | Protein coding | - | E9PEI4 | - | TSL:1, GENCODE basic, |
| PTK2-246 | ENST00000523539.5 | 3236 | 724aa | Protein coding | - | H0YB16 | - | CDS 5' incomplete, TSL:2, |
| PTK2-238 | ENST00000521986.5 | 2393 | 753aa | Protein coding | - | H0YBZ1 | - | CDS 5' incomplete, TSL:2, |
| PTK2-216 | ENST00000519881.5 | 863 | 121aa | Protein coding | - | E5RFW9 | - | TSL:3, GENCODE basic, |
| PTK2-252 | ENST00000523803.5 | 750 | 120aa | Protein coding | - | E5RI72 | - | CDS 3' incomplete, TSL:3, |
| PTK2-233 | ENST00000521562.5 | 678 | 132aa | Protein coding | - | E5RJQ2 | - | CDS 3' incomplete, TSL:4, |
| PTK2-231 | ENST00000521332.5 | 630 | 65aa | Protein coding | - | E5RIK4 | - | CDS 3' incomplete, TSL:4, |
| PTK2-244 | ENST00000523435.1 | 629 | 120aa | Protein coding | - | E5RI03 | - | CDS 3' incomplete, TSL:4, |
| PTK2-219 | ENST00000520045.5 | 602 | 117aa | Protein coding | - | E5RH01 | - | CDS 3' incomplete, TSL:4, |
| PTK2-243 | ENST00000523388.5 | 581 | 67aa | Protein coding | - | E5RHK7 | - | CDS 3' incomplete, TSL:2, |
| PTK2-210 | ENST00000519024.5 | 576 | 168aa | Protein coding | - | E5RJI4 | - | CDS 3' incomplete, TSL:4, |
| PTK2-223 | ENST00000520828.5 | 574 | 59aa | Protein coding | - | E5RG86 | - | CDS 3' incomplete, TSL:4, |
| PTK2-220 | ENST00000520151.5 | 573 | 124aa | Protein coding | - | E5RI29 | - | TSL:4, GENCODE basic, |
| PTK2-205 | ENST00000517453.5 | 571 | 117aa | Protein coding | - | E5RH01 | - | CDS 3' incomplete, TSL:4, |
| PTK2-225 | ENST00000520892.5 | 571 | 121aa | Protein coding | - | E5RFW9 | - | TSL:4, GENCODE basic, |
| PTK2-222 | ENST00000520475.5 | 570 | 156aa | Protein coding | - | E5RGA6 | - | CDS 3' incomplete, TSL:5, |
| PTK2-239 | ENST00000522424.5 | 569 | 133aa | Protein coding | - | E5RG80 | - | CDS 3' incomplete, TSL:4, |
| PTK2-241 | ENST00000522950.1 | 548 | 67aa | Protein coding | - | E5RII9 | - | TSL:4, GENCODE basic, |
| PTK2-235 | ENST00000521907.5 | 542 | 119aa | Protein coding | - | E5RG66 | - | CDS 3' incomplete, TSL:4, |
| PTK2-232 | ENST00000521395.5 | 540 | 107aa | Protein coding | - | E5RH48 | - | CDS 3' incomplete, TSL:4, |
| PTK2-257 | ENST00000524357.5 | 535 | 122aa | Protein coding | - | E5RJP0 | - | CDS 3' incomplete, TSL:4, |
| PTK2-249 | ENST00000523679.1 | 504 | 14aa | Protein coding | - | E5RJN1 | - | CDS 3' incomplete, TSL:3, |
| PTK2-254 | ENST00000524040.5 | 475 | 51aa | Protein coding | - | E5RH08 | - | CDS 3' incomplete, TSL:2, |
| PTK2-256 | ENST00000524257.5 | 430 | 8aa | Protein coding | - | A0A1D5RMT1 | - | CDS 3' incomplete, TSL:4, |
| PTK2-218 | ENST00000519993.5 | 4388 | 66aa | Nonsense mediated decay | - | E5RG54 | - | TSL:1, |
| PTK2-255 | ENST00000524202.5 | 3014 | 319aa | Nonsense mediated decay | - | H0YB99 | - | CDS 5' incomplete, TSL:1, |
| PTK2-230 | ENST00000521250.5 | 592 | 72aa | Nonsense mediated decay | - | H0YAS0 | - | CDS 5' incomplete, TSL:4, |
| PTK2-227 | ENST00000521029.5 | 584 | 79aa | Nonsense mediated decay | - | E5RGP1 | - | TSL:4, |
| PTK2-234 | ENST00000521791.5 | 556 | 66aa | Nonsense mediated decay | - | E5RG54 | - | TSL:4, |
| PTK2-236 | ENST00000521981.5 | 552 | 128aa | Nonsense mediated decay | - | H0YB33 | - | CDS 5' incomplete, TSL:5, |
| PTK2-247 | ENST00000523670.5 | 542 | 72aa | Nonsense mediated decay | - | E5RIR5 | - | TSL:4, |
| PTK2-242 | ENST00000523067.5 | 488 | 66aa | Nonsense mediated decay | - | E5RG54 | - | TSL:4, |
| PTK2-214 | ENST00000519635.1 | 578 | No protein | Processed transcript | - | - | - | TSL:4, |
| PTK2-250 | ENST00000523746.5 | 574 | No protein | Processed transcript | - | - | - | TSL:4, |
| PTK2-221 | ENST00000520460.1 | 572 | No protein | Processed transcript | - | - | - | TSL:4, |
| PTK2-209 | ENST00000518509.5 | 569 | No protein | Processed transcript | - | - | - | TSL:4, |
| PTK2-245 | ENST00000523474.5 | 567 | No protein | Processed transcript | - | - | - | TSL:5, |
| PTK2-211 | ENST00000519361.5 | 553 | No protein | Processed transcript | - | - | - | TSL:5, |
| PTK2-237 | ENST00000521985.1 | 538 | No protein | Processed transcript | - | - | - | TSL:3, |
| PTK2-229 | ENST00000521172.1 | 507 | No protein | Processed transcript | - | - | - | TSL:5, |
| PTK2-253 | ENST00000523805.5 | 440 | No protein | Processed transcript | - | - | - | TSL:3, |
| PTK2-224 | ENST00000520843.5 | 438 | No protein | Processed transcript | - | - | - | TSL:5, |
| PTK2-251 | ENST00000523797.1 | 405 | No protein | Processed transcript | - | - | - | TSL:3, |
| PTK2-217 | ENST00000519899.5 | 762 | No protein | Retained intron | - | - | - | TSL:2, |
| PTK2-226 | ENST00000520917.1 | 653 | No protein | Retained intron | - | - | - | TSL:4, |
| PTK2-248 | ENST00000523675.1 | 650 | No protein | Retained intron | - | - | - | TSL:3, |
| PTK2-204 | ENST00000510126.2 | 580 | No protein | Retained intron | - | - | - | TSL:4, |
| PTK2-208 | ENST00000518173.5 | 441 | No protein | Retained intron | - | - | - | TSL:2, |




Red Cotton™ Assessment
Project Difficulty Level unknown
| Target Gene | PTK2 |
| This KO Strategy | loading |
| Red Cotton™ Notes | Gene PTK2 had been KO in hek293t cell line. |
Aforementioned information comes from Ubigene database. Different origin of cell lines may have
different condition. Ubigene reserved all the right for final explanation.
Special deals for this gene:
$49
Single gRNA plasmid off-shelf
$599
Single gRNA lentivirus



Work flow
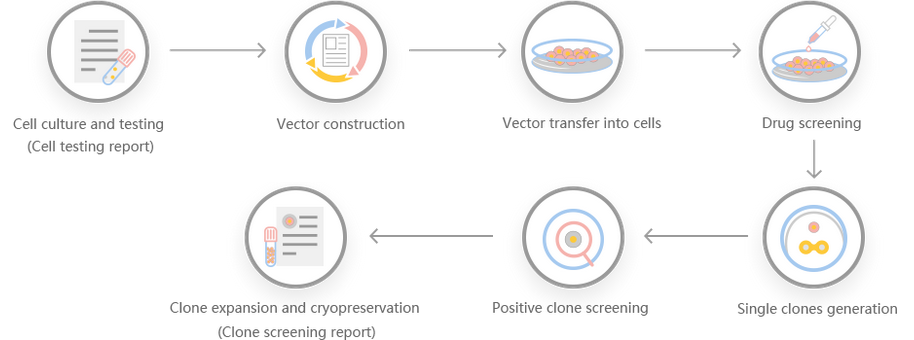






 Comment:
Comment: