CRISPR-U™ Gene Knockout Cell Line
Strategy
AKT1 Gene Knockout Strategy
CRISPR-U™ technology (CRISPR based), developed by Ubigene, is more efficient than general
CRISPR/Cas9 technology in double-strand breaking and homologous recombination. With CRISPR-U™, Ubigene has
successfully edited over 3000 genes on more than 100 types of cell lines.
Objective
To create a Human AKT1 Knockout
model in cell line by
CRISPR-U™-mediated
genome
engineering.
Target gene info 
| Official symbol | AKT1 |
| Gene id | 207 |
| Organism | Homo sapiens |
| Official full symbol | AKT serine/threonine kinase 1 |
| Gene type | protein-coding |
| Also known as | AKT, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA |
| Summary | This gene encodes one of the three members of the human AKT serine-threonine protein kinase family which are often referred to as protein kinase B alpha, beta, and gamma. These highly similar AKT proteins all have an N-terminal pleckstrin homology domain, a serine/threonine-specific kinase domain and a C-terminal regulatory domain. These proteins are phosphorylated by phosphoinositide 3-kinase (PI3K). AKT/PI3K forms a key component of many signalling pathways that involve the binding of membrane-bound ligands such as receptor tyrosine kinases, G-protein coupled receptors, and integrin-linked kinase. These AKT proteins therefore regulate a wide variety of cellular functions including cell proliferation, survival, metabolism, and angiogenesis in both normal and malignant cells. AKT proteins are recruited to the cell membrane by phosphatidylinositol 3,4,5-trisphosphate (PIP3) after phosphorylation of phosphatidylinositol 4,5-bisphosphate (PIP2) by PI3K. Subsequent phosphorylation of both threonine residue 308 and serine residue 473 is required for full activation of the AKT1 protein encoded by this gene. Phosphorylation of additional residues also occurs, for example, in response to insulin growth factor-1 and epidermal growth factor. Protein phosphatases act as negative regulators of AKT proteins by dephosphorylating AKT or PIP3. The PI3K/AKT signalling pathway is crucial for tumor cell survival. Survival factors can suppress apoptosis in a transcription-independent manner by activating AKT1 which then phosphorylates and inactivates components of the apoptotic machinery. AKT proteins also participate in the mammalian target of rapamycin (mTOR) signalling pathway which controls the assembly of the eukaryotic translation initiation factor 4F (eIF4E) complex and this pathway, in addition to responding to extracellular signals from growth factors and cytokines, is disregulated in many cancers. Mutations in this gene are associated with multiple types of cancer and excessive tissue growth including Proteus syndrome and Cowden syndrome 6, and breast, colorectal, and ovarian cancers. Multiple alternatively spliced transcript variants have been found for this gene. |
| Genomic regions | Chromosome 14 |
Strategy Summary
This gene has 9 protein coding transcripts:
| Name | Transcript ID | bp | Protein | Biotype | CCDS | UniProt Match | RefSeq Match | Flags |
| AKT1-208 | ENST00000554581.5 | 3916 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-202 | ENST00000402615.6 | 3913 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-214 | ENST00000555528.5 | 3066 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-219 | ENST00000649815.2 | 2957 | 480aa | Protein coding | CCDS9994 | P31749-1 | NM_001382430.1 | GENCODE basic, APPRIS P1, MANE Select v0.92, |
| AKT1-201 | ENST00000349310.7 | 2866 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-203 | ENST00000407796.7 | 2779 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-211 | ENST00000554848.5 | 1595 | 480aa | Protein coding | CCDS9994 | P31749-1 | - | TSL:1, GENCODE basic, APPRIS P1, |
| AKT1-207 | ENST00000554192.5 | 922 | 184aa | Protein coding | - | G3V2I6 | - | CDS 5' incomplete, TSL:2, |
| AKT1-213 | ENST00000555458.5 | 705 | 170aa | Protein coding | - | G3V3X1 | - | CDS 5' incomplete, TSL:2, |
| AKT1-209 | ENST00000554585.5 | 858 | 146aa | Nonsense mediated decay | - | A0A087WY56 | - | CDS 5' incomplete, TSL:3, |
| AKT1-204 | ENST00000544168.5 | 1564 | No protein | Processed transcript | - | - | - | TSL:2, |
| AKT1-206 | ENST00000553797.1 | 783 | No protein | Processed transcript | - | - | - | TSL:3, |
| AKT1-212 | ENST00000555380.1 | 534 | No protein | Processed transcript | - | - | - | TSL:4, |
| AKT1-216 | ENST00000557494.1 | 408 | No protein | Processed transcript | - | - | - | TSL:2, |
| AKT1-217 | ENST00000557552.1 | 8396 | No protein | Retained intron | - | - | - | TSL:5, |
| AKT1-218 | ENST00000610370.1 | 3187 | No protein | Retained intron | - | - | - | TSL:2, |
| AKT1-205 | ENST00000553506.5 | 2830 | No protein | Retained intron | - | - | - | TSL:2, |
| AKT1-215 | ENST00000556836.1 | 592 | No protein | Retained intron | - | - | - | TSL:4, |
| AKT1-210 | ENST00000554826.1 | 572 | No protein | Retained intron | - | - | - | TSL:3, |




Red Cotton™ Assessment
Project Difficulty Level unknown
| Target Gene | AKT1 |
| This KO Strategy | loading |
| Red Cotton™ Notes | Gene AKT1 had been KO in hela cell line. |
Aforementioned information comes from Ubigene database. Different origin of cell lines may have
different condition. Ubigene reserved all the right for final explanation.
Special deals for this gene:
$49
Single gRNA plasmid off-shelf
$599
Single gRNA lentivirus



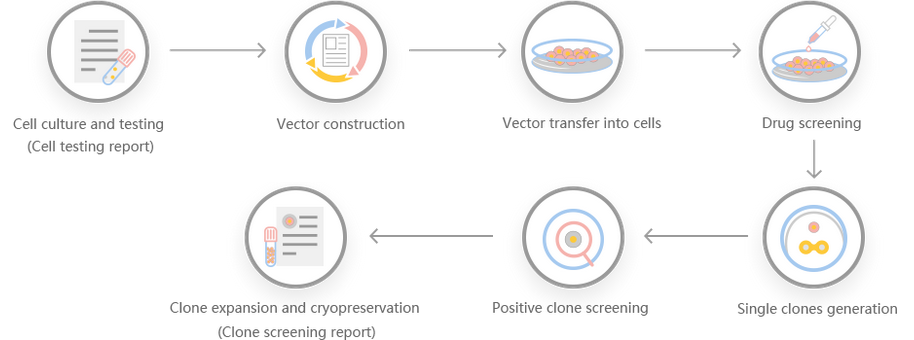
Work flow






 Comment:
Comment: